Modeling antibodies
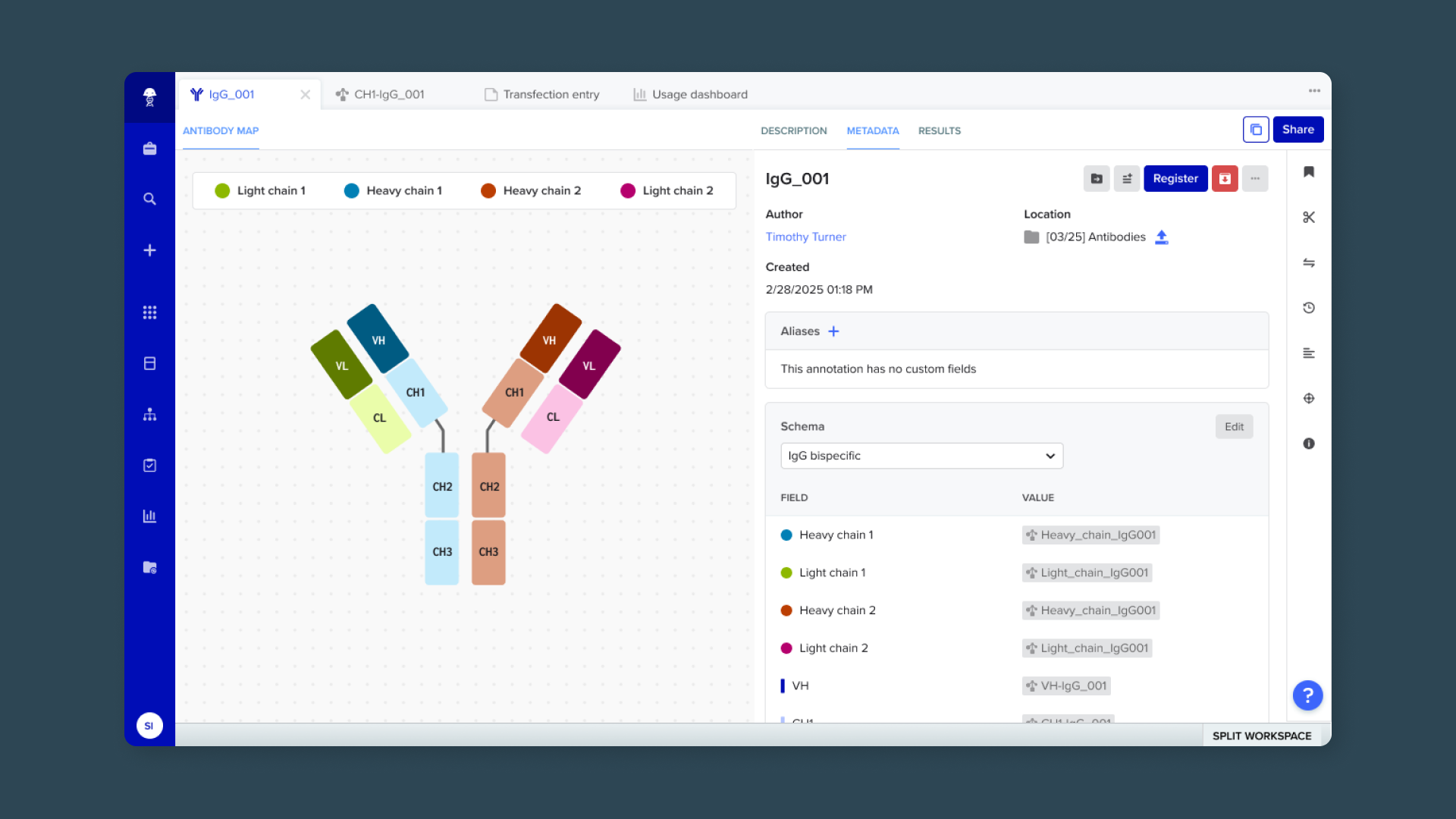
Overview
Antibody discovery is one of the fastest-growing areas in biologics R&D, and customers rely on Benchling to model the complexity of antibody structures and relationships at scale. Each antibody is composed of nested biological components—chains, domains, DNA sequences, and plasmids—that scientists must register, link, and analyze to trace molecular lineage and performance.
This project unifies multiple product efforts under a single goal: enabling users to create, model, and visualize antibodies and their component sequences. My role spans end-to-end design for data ingestion, modeling, and visualization flows, in partnership with product and engineering leads across three scrum teams.
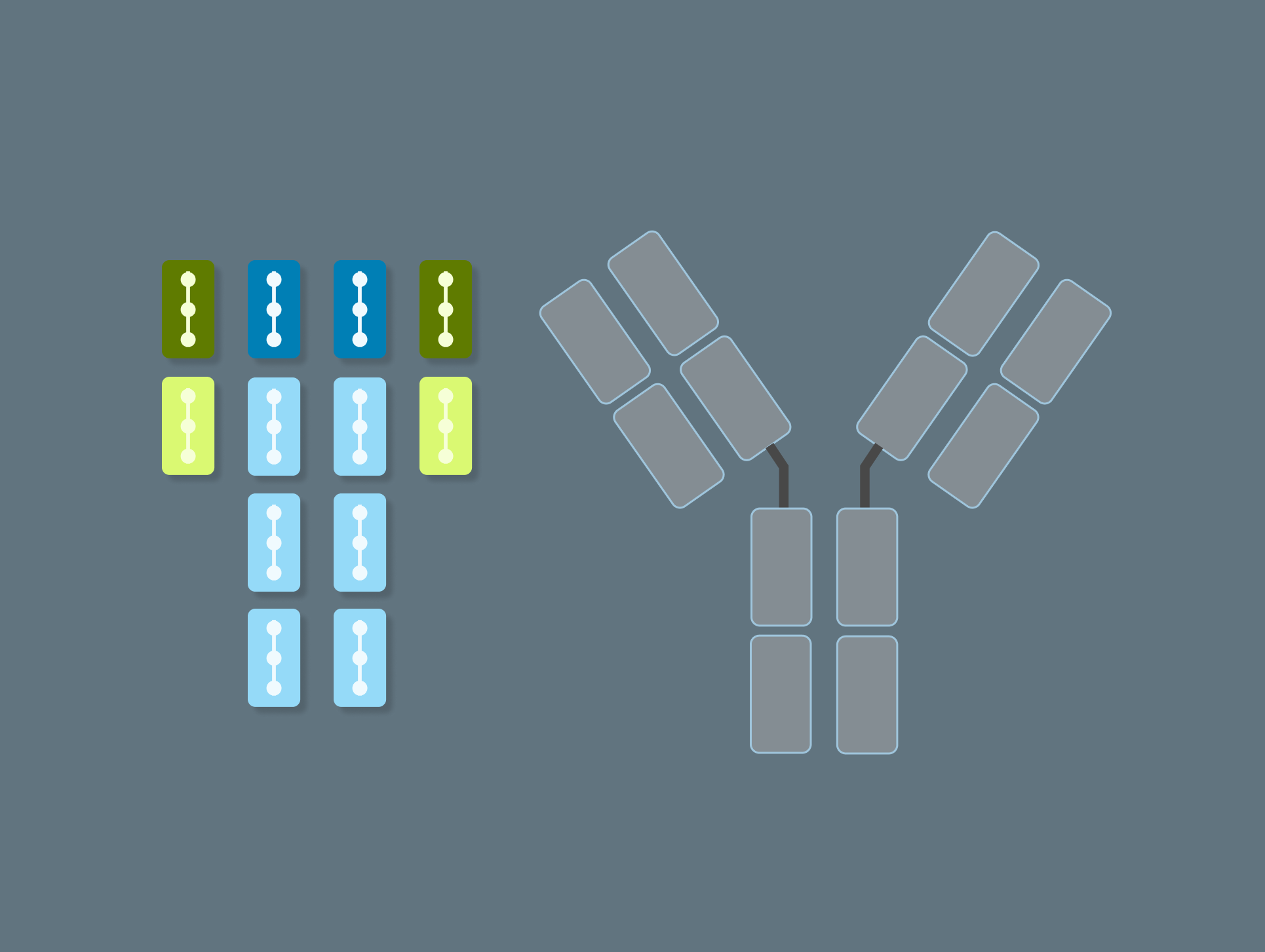
Model antibodies and their components
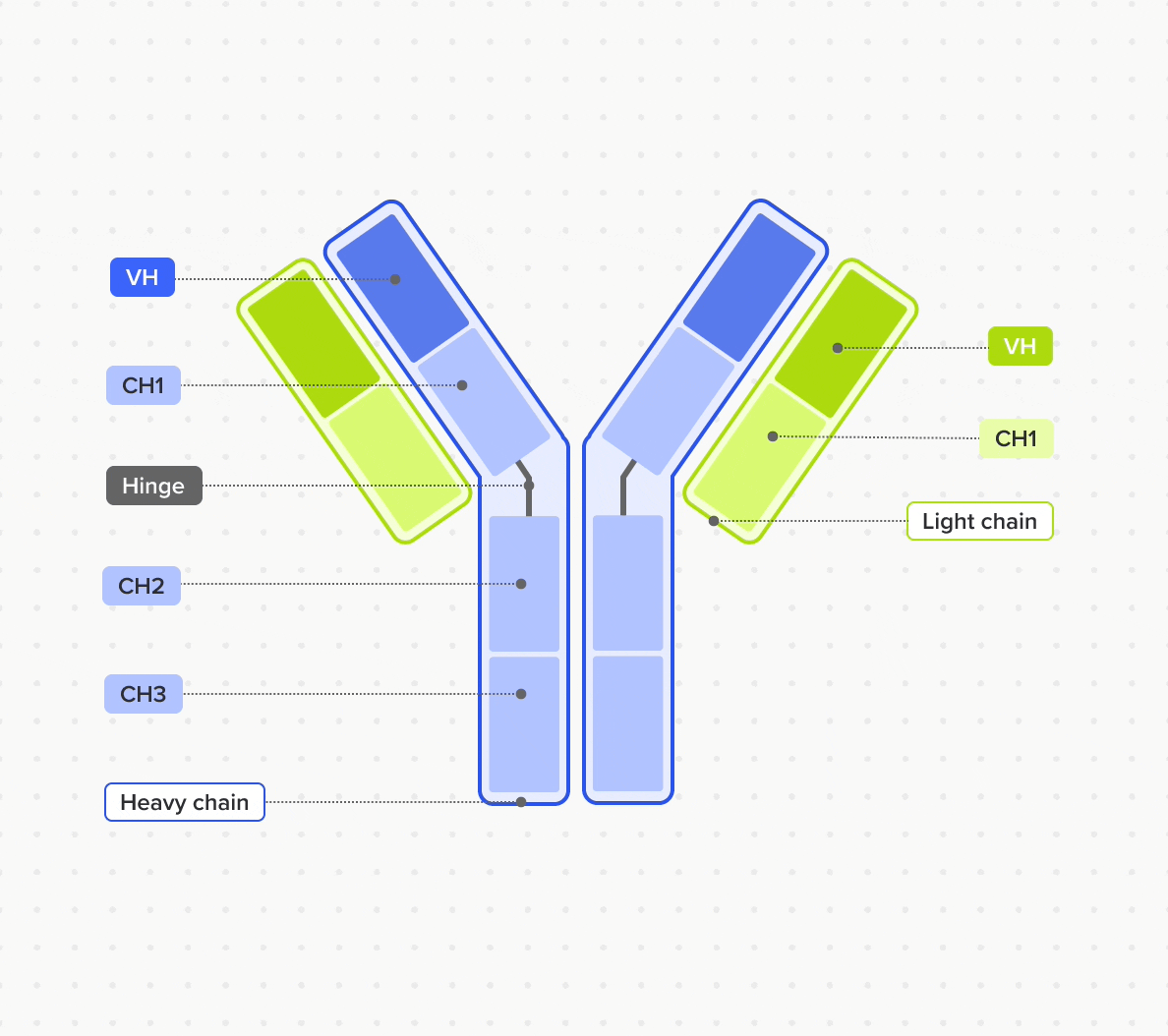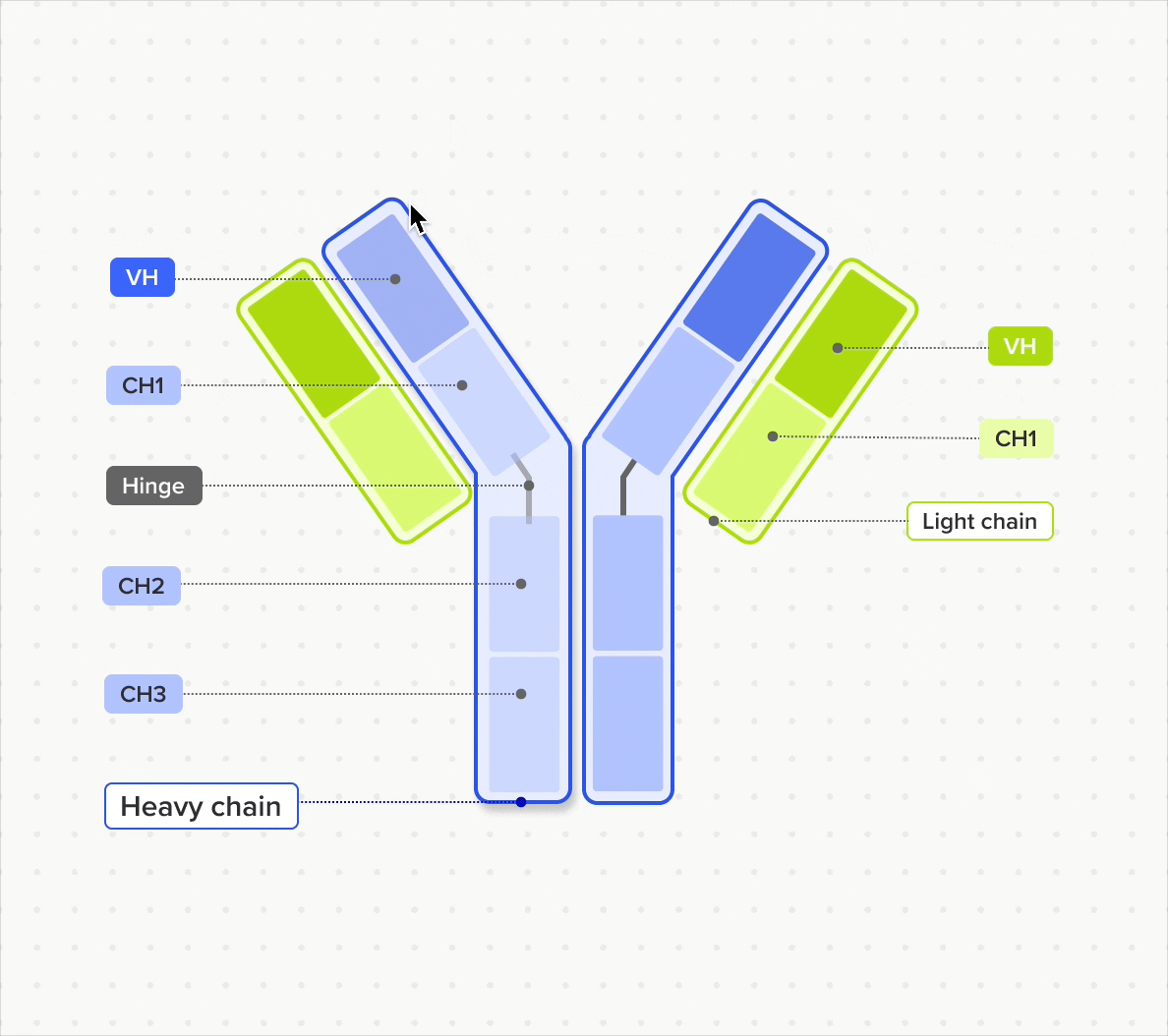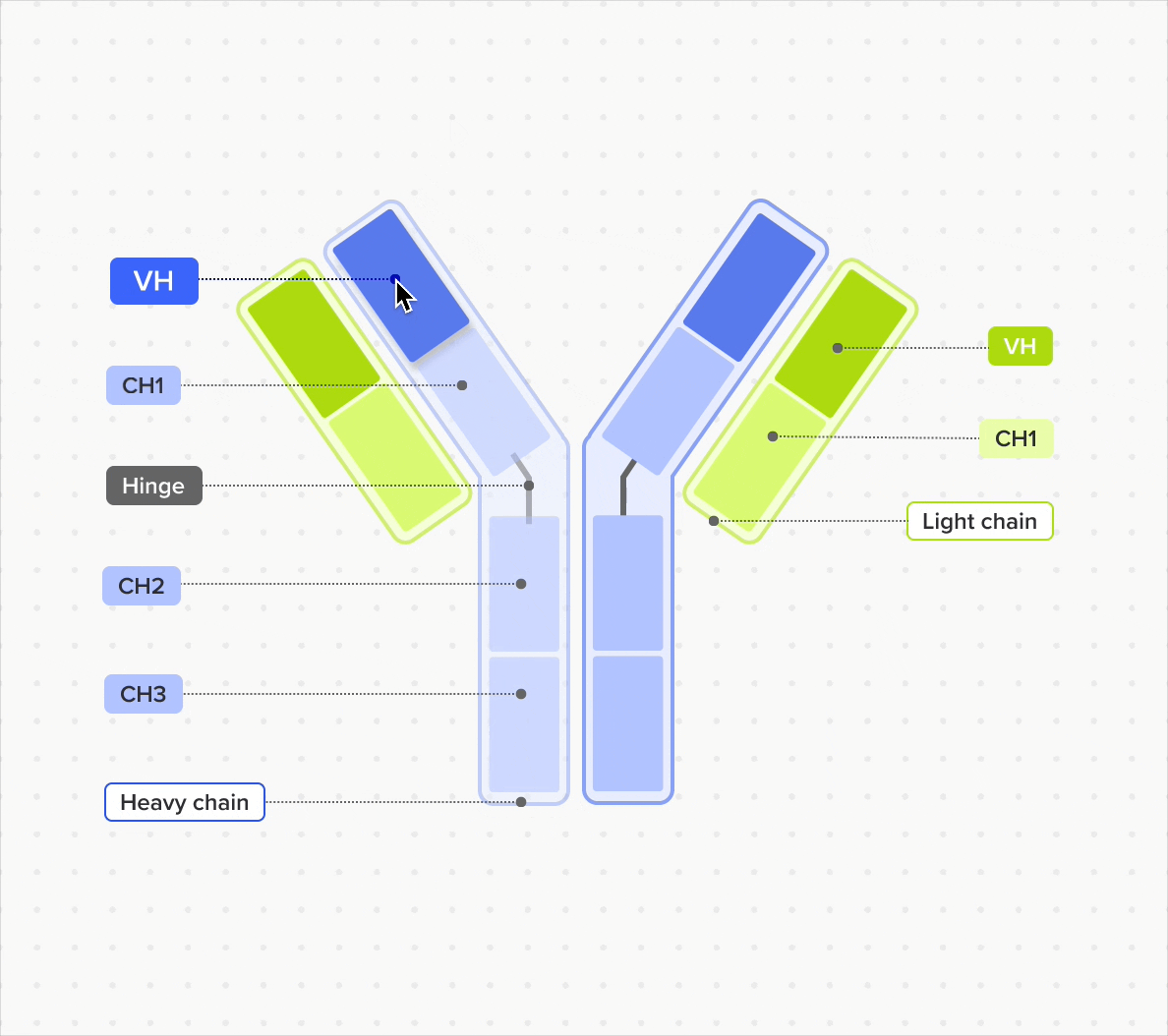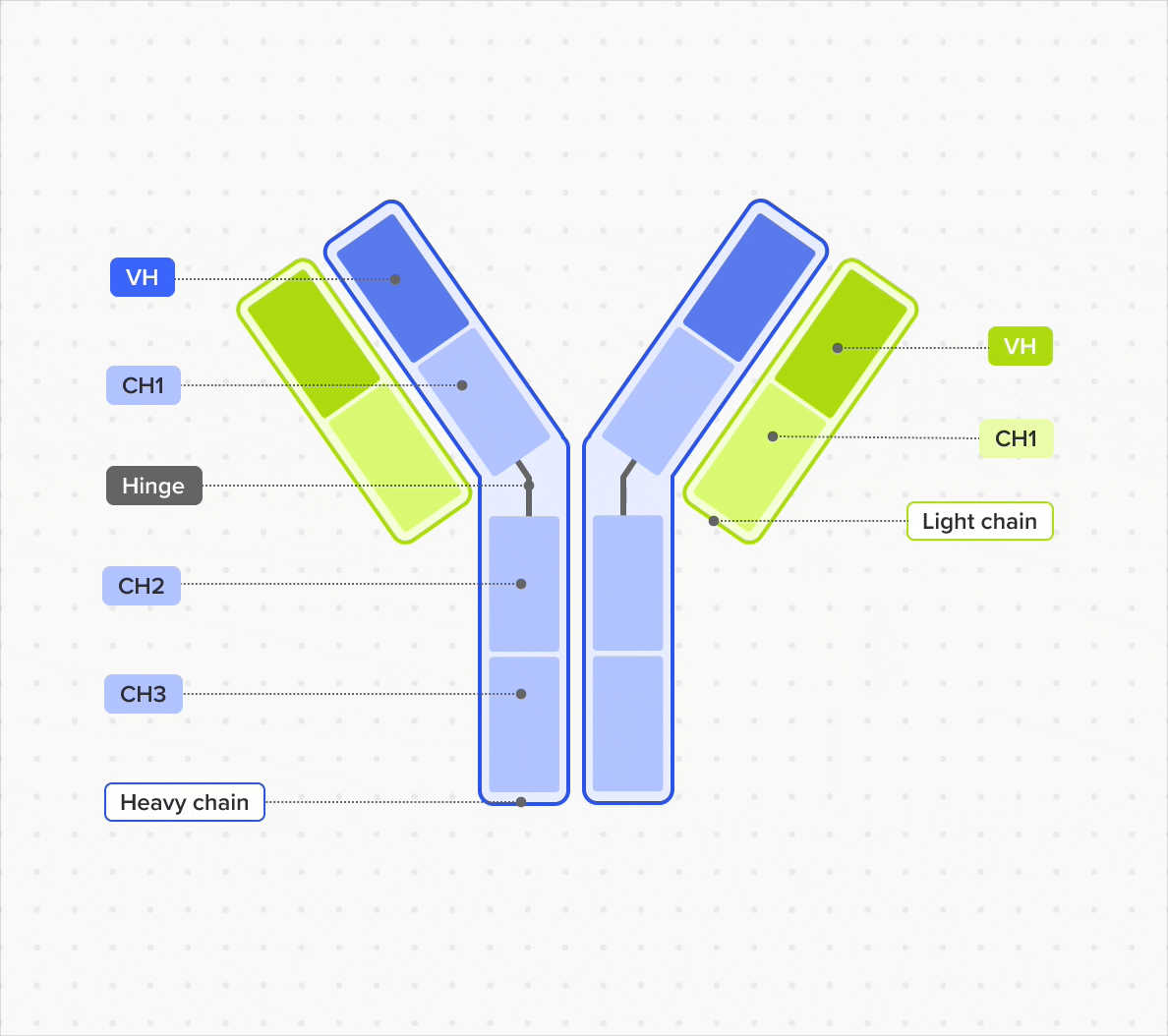
Design and represent complete antibody structures from chains and domains across both DNA and amino acid sequences, all linked within a unified data model
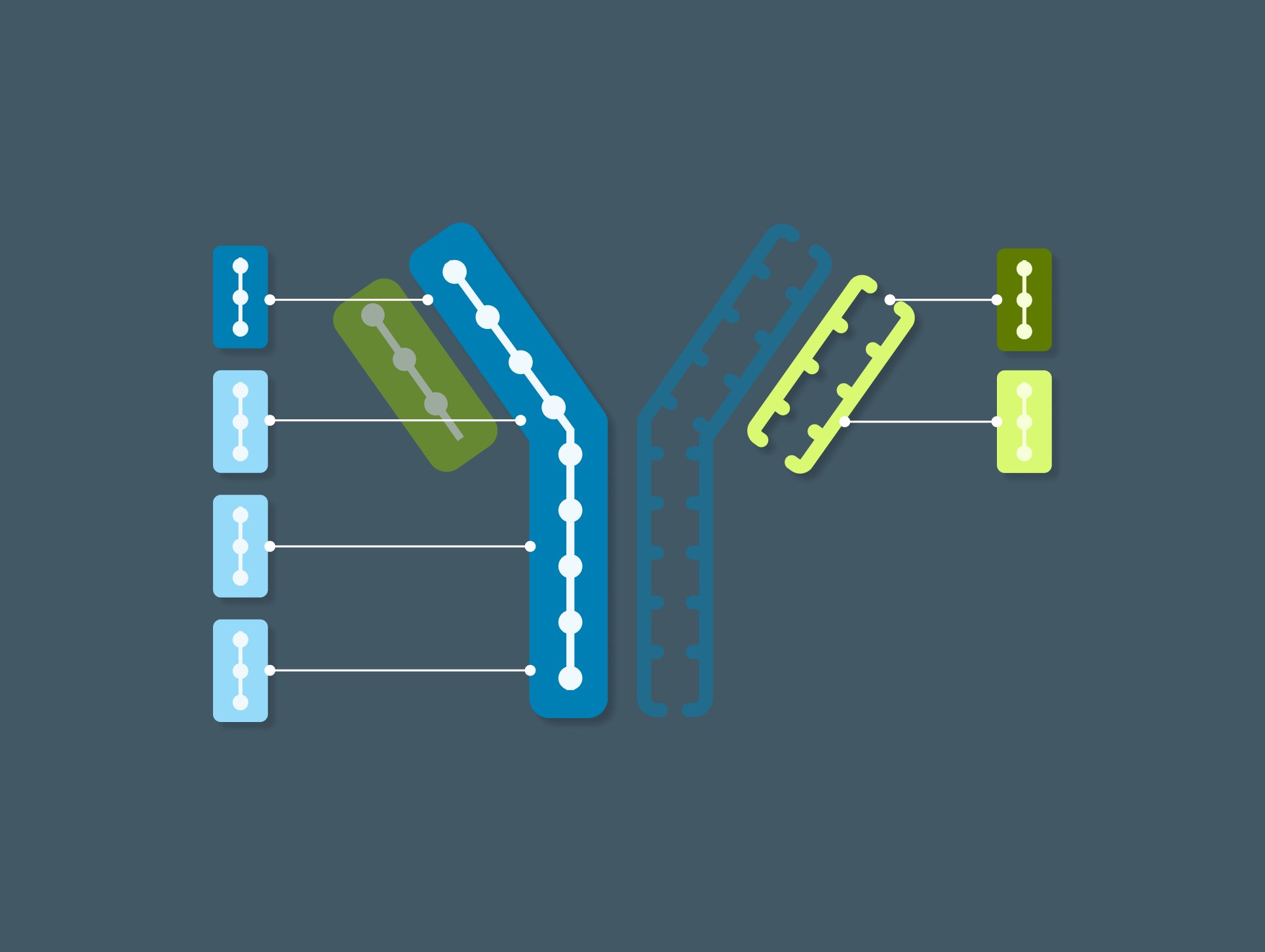
Maintain lineage and traceability
Link domains to chains and chains to antibodies to navigate the full molecular hierarchy of the antibody data model and maintain complete lineage and contextual traceability
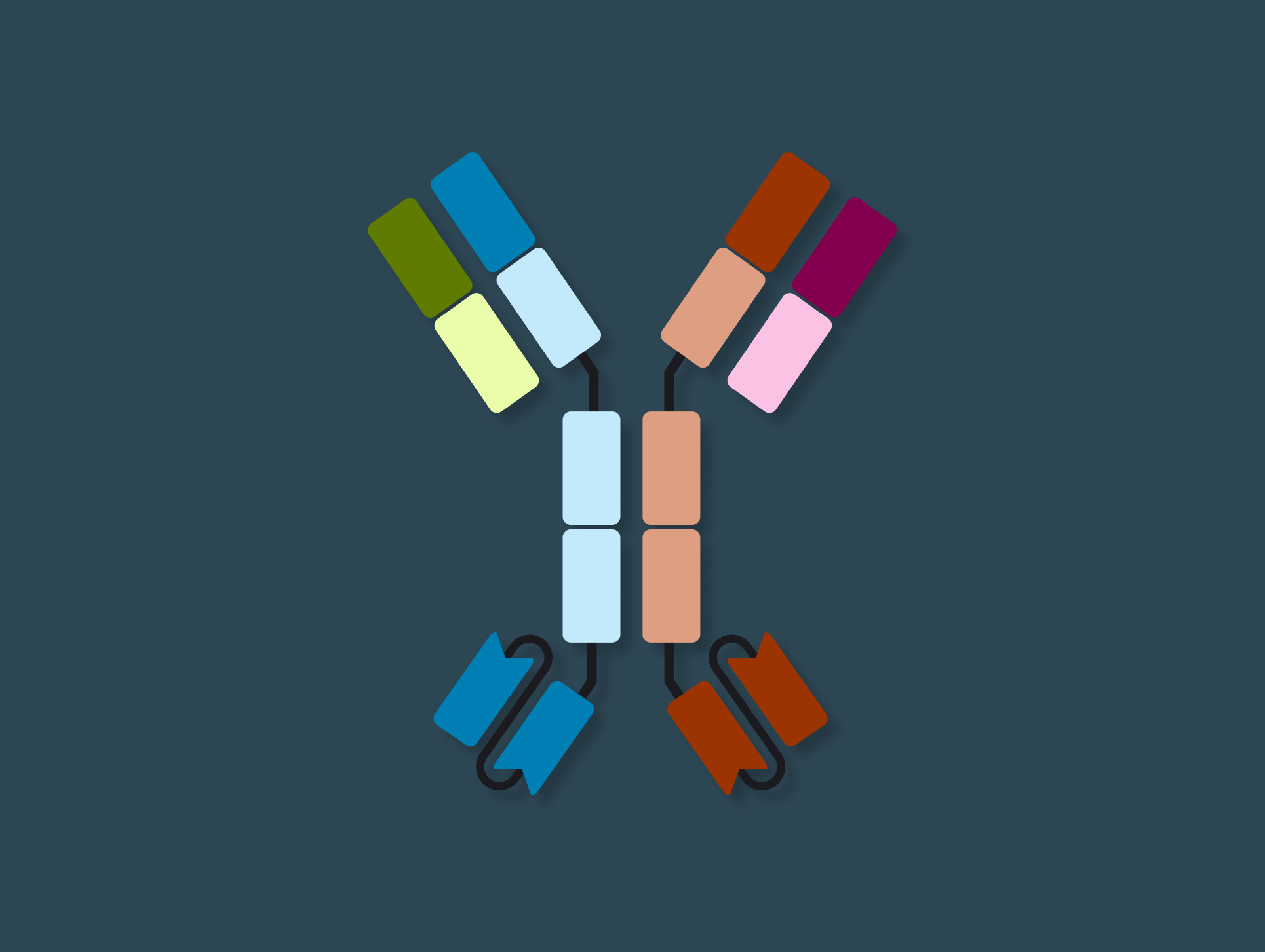
Support diverse antibody formats
Model and support antibodies of diverse specificities, subtypes, and isotypes through flexible, customizable tools that adapt to any antibody format
Problem
Fragmented and manual antibody modeling
Before this work, antibody registration and modeling were fragmented and highly manual:
- Scientists had to create every entity individually (DNA, amino acid sequences, chains, and antibodies), then manually link them through schemas.
- Admins and informatics teams couldn’t represent their organization’s unique antibody formats without engineering help, limiting adoption and data completeness.
- Benchling’s existing link systems (“biointelligent links”) were inconsistent and didn’t validate biological relationships, creating data integrity and usability risks
As a result, scientists and admins alike struggled to capture the full picture of their antibody research, losing time and confidence in data that should have accelerated discovery.
Use cases
Three primary use cases to support antibody workflows:
High-throughput data ingestion - scientists want to create hundreds of antibodies at a time, starting with DNA and amino acid sequences, and have Benchling automatically create linked entities—chains, domains, and antibodies—within a single workflow.
Modeling and linking entities intelligently - biointelligent links should automatically detect and validate relationships like translation (DNA → AA) and part (domain → chain), so that scientists can trace biological lineage without manual linking
Custom antibody formats for enterprise admins - admins need tools to define new antibody formats from building blocks (domains, chains, linkers, hinges). This allows each organization to model proprietary constructs unique to their IP, without engineering support


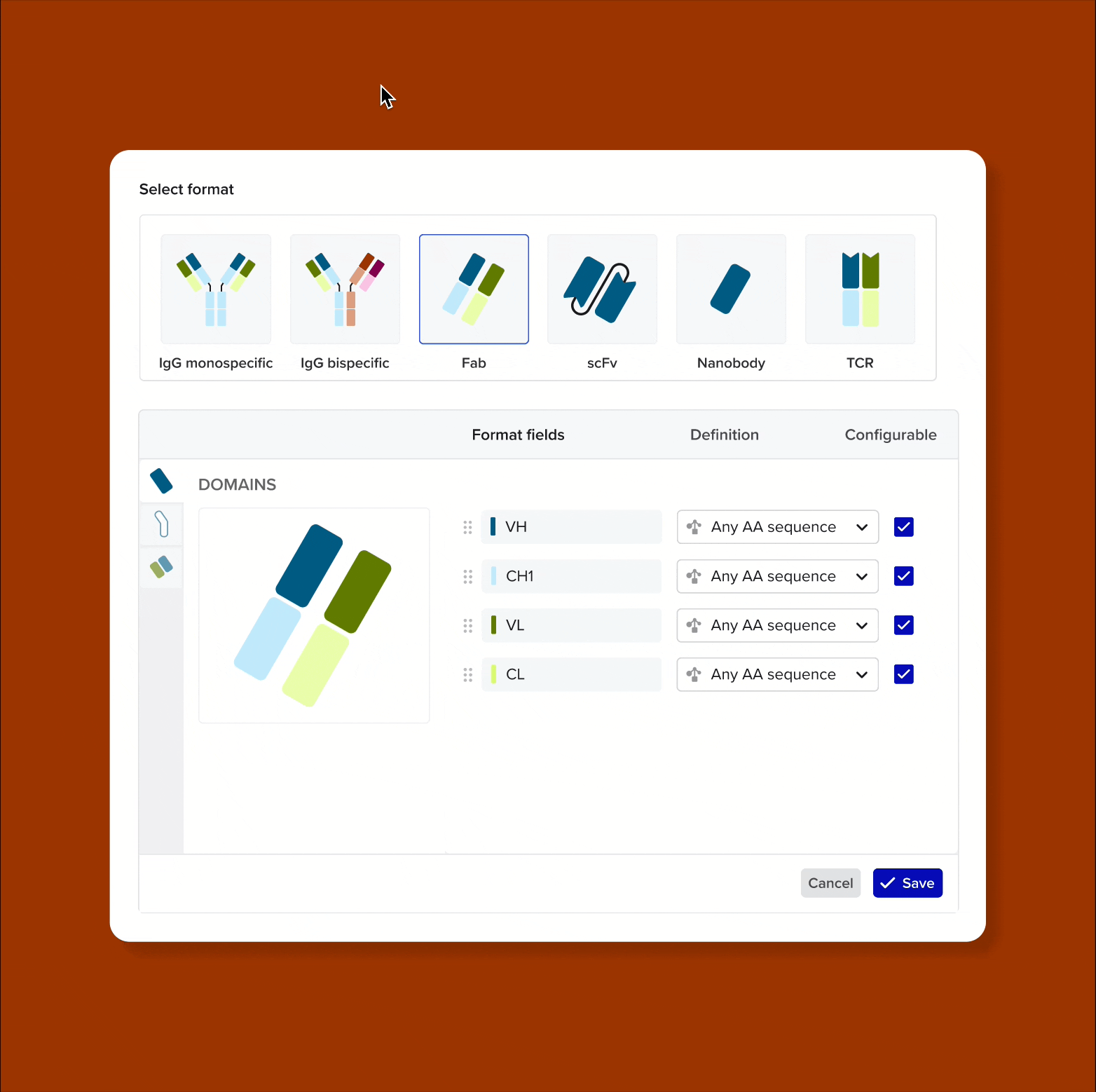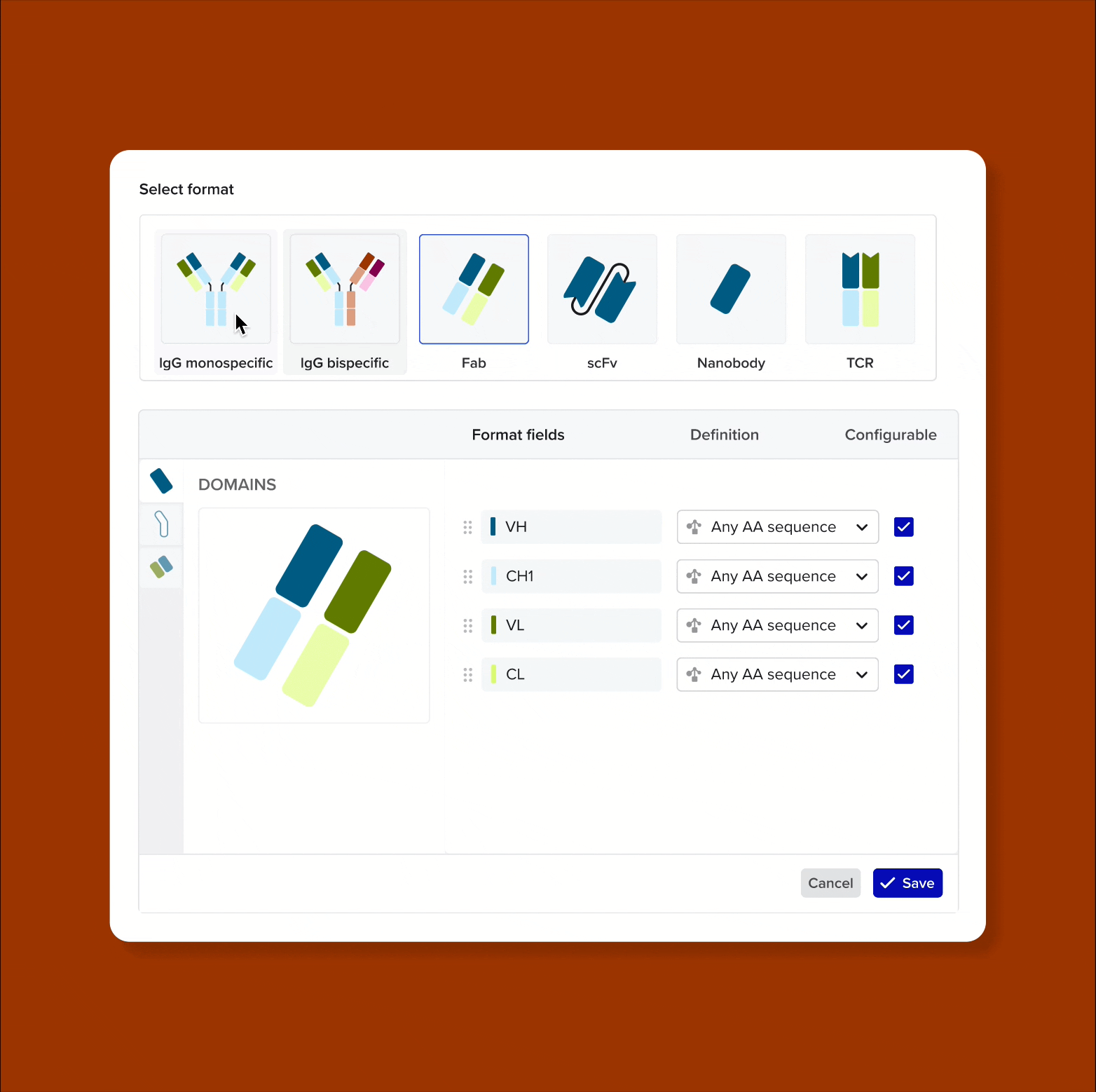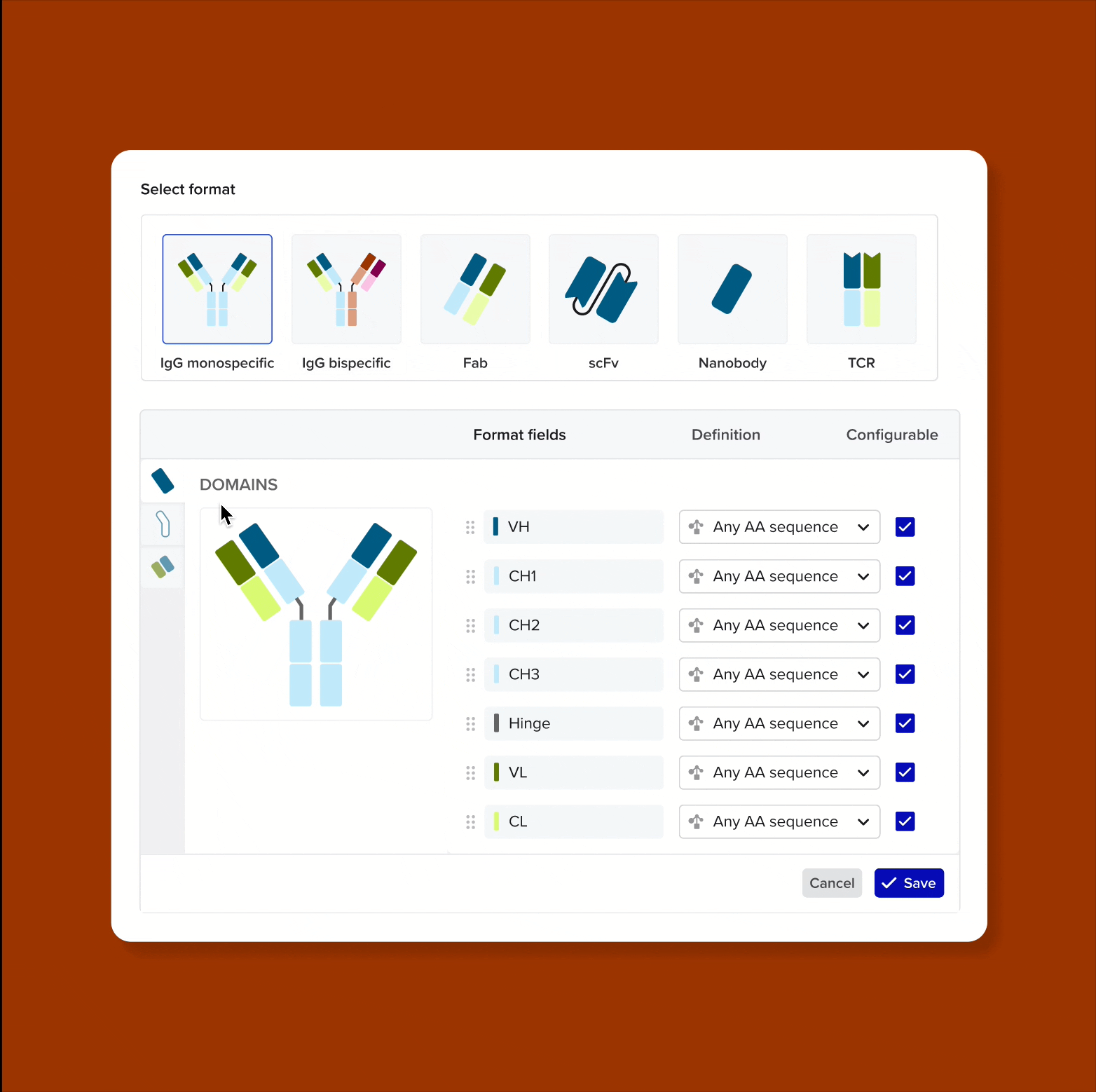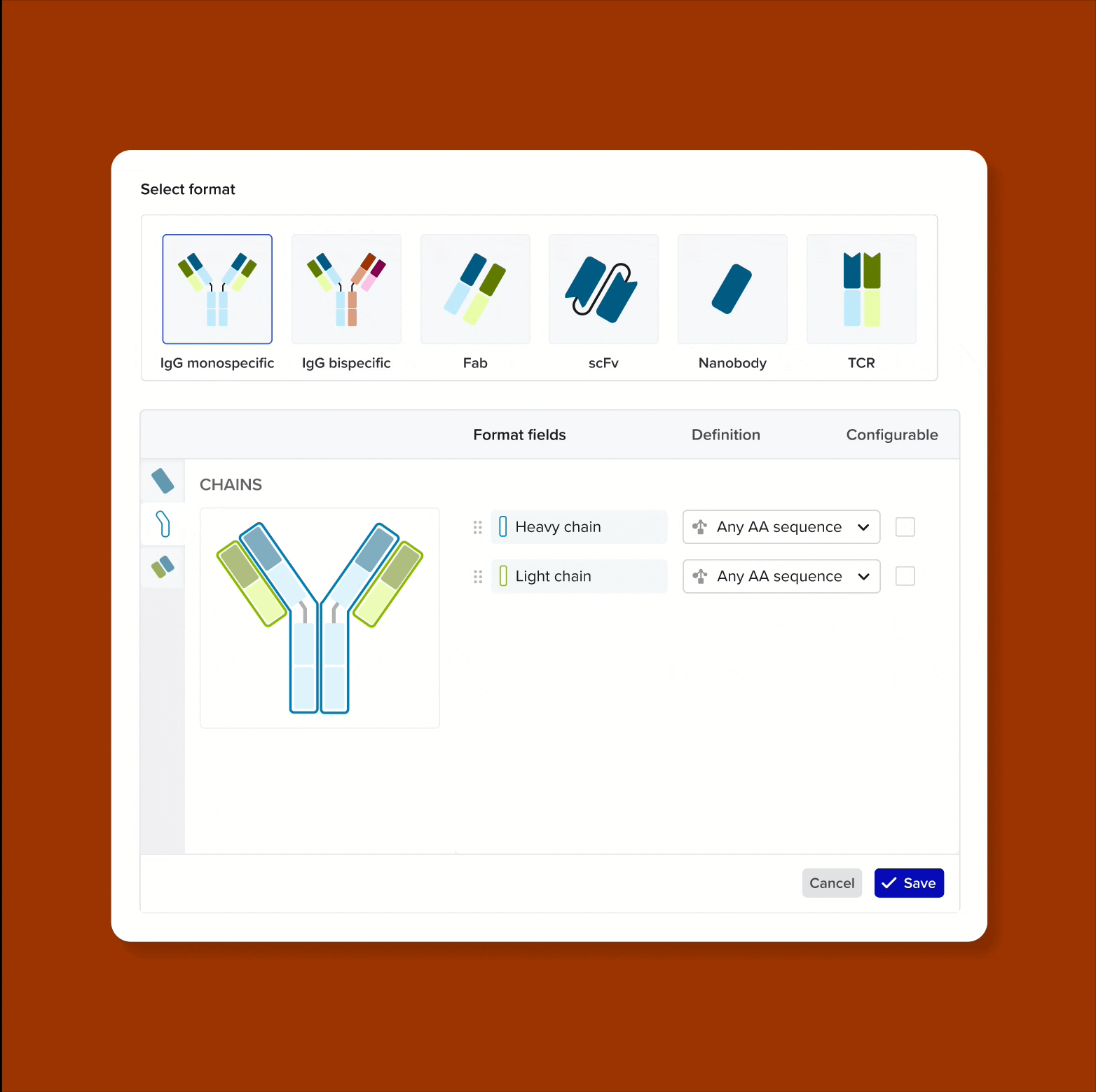
Solution
This project is a WIP, and I'm currently grinding to deliver a best-in-class MVP solution. Stay tuned for updates!









